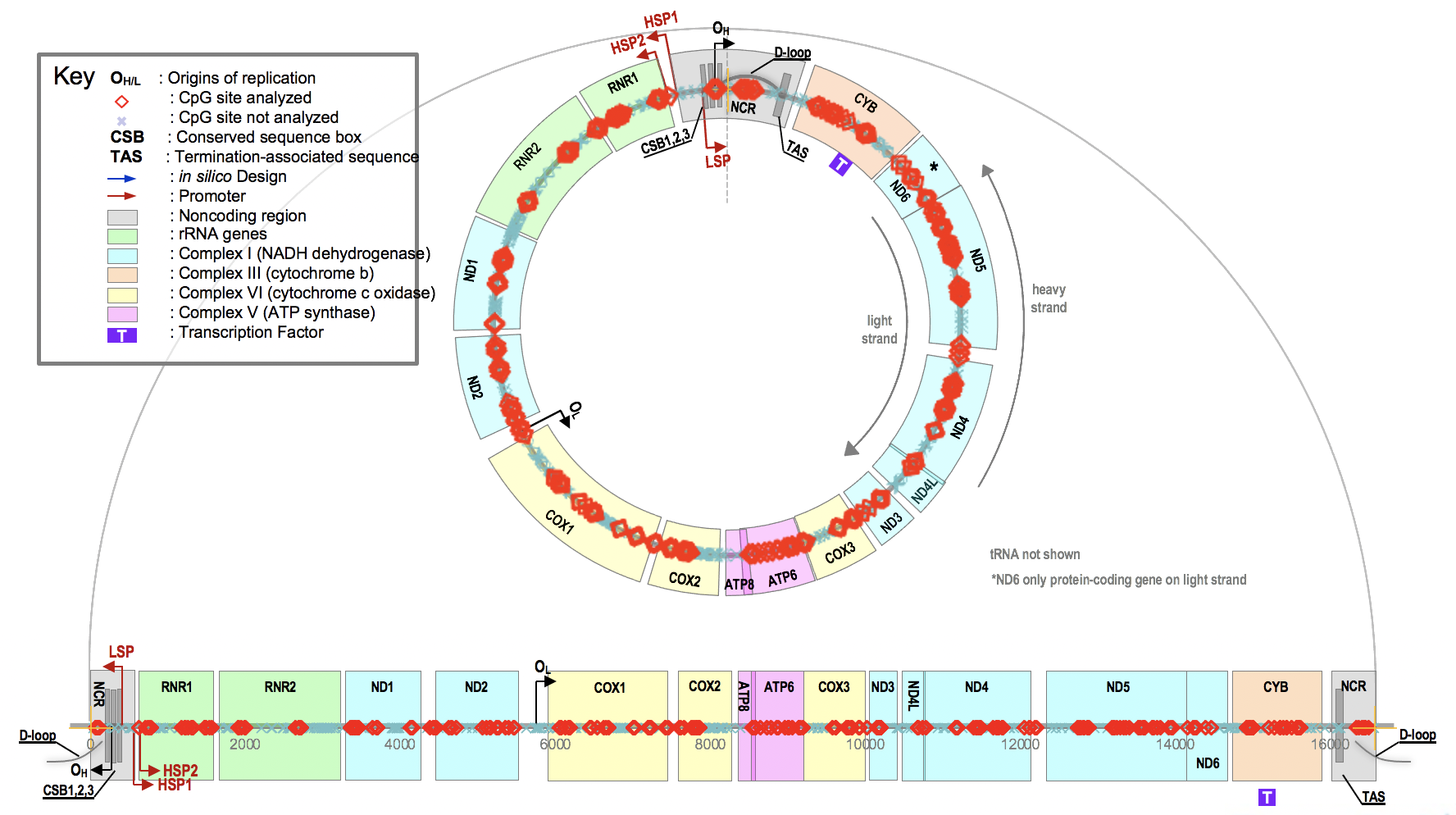
Mitochondria are governed by a symbiotic relationship between two genomes: the mitochondrial and the nuclear DNA. As a result, changes in nuclear gene expression caused by mutations and epigenetic modifications can have an impact on mitochondrial function. The Mitochondrial Methylation Panel (N109V0) is a targeted resequencing assay that uses multiplex PCR to evaluate the human mitochondrial genome for alterations. The panel provides coverage of 11 mitochondrial genes by 52 individual PCR in the ~16.569 kb mitochondrial genome, enabling for the detection of significant variants.
Other DNA Methylation Panels
Figure 1. Human Mitochondrial Genome NCBI Reference Sequence: NC_012920.1
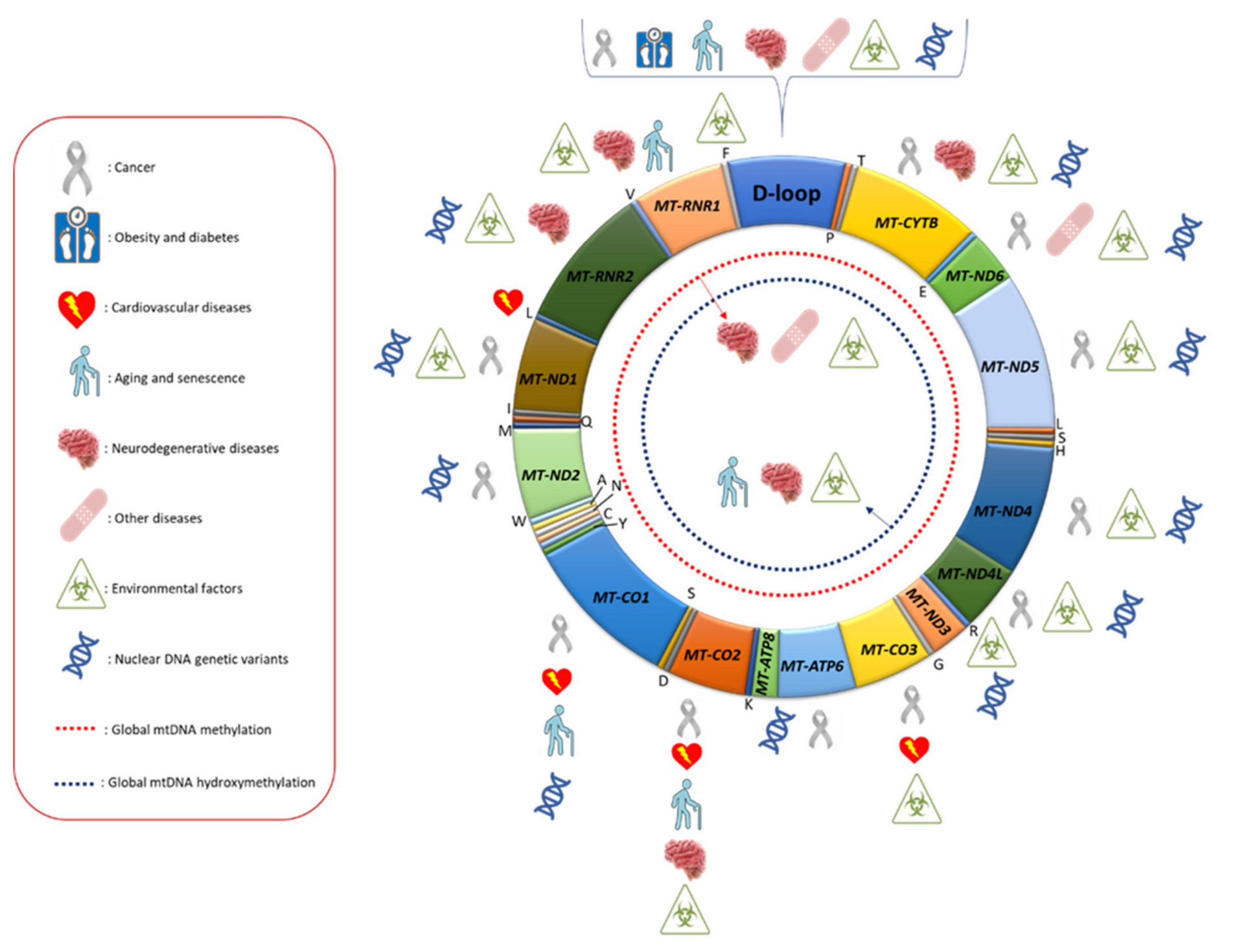
Figure 2. Mitochondrial DNA regions whose methylation levels were found to be associated with different human diseases, environmental factors and nuclear DNA genetic variants. In the red box, the meaning of the symbols used is reported.
- Complete Solution
Detect and analyze only the regions and genes of the human mitochondrial genome using Targeted Next-Gen Bisulfite Sequencing - Economical & Faster Turnaround-Time
Perform the DNA extraction, bisulfite modification, library preparation, high throughput sequencing, data analysis, and assay validation - Exceptional Work
Produce higher quality data due to significantly greater coverage of each CpG site
| Parameter | Specification |
| Method |
|
| Sequencing Platforms | Illumina®, Ion Torrent™ |
| Number of Genese | 11 by individual PCR of ~16.569 kb |
| Number of Amplicons | 52 amplicons and ~40 amplicon has >100 x coverage in NGS |
| No. of CpGs | 243 CpGs |
| Size (bp) | 8092 bp |
| Design Coverage | ~200 CpG sites with >100x in gNGS |
| Input DNA Requirement | 250 extracted DNA |
| No. of Primer Sets | 52 individual PCR sets |
| No. of Reactions | 8 multiplex PCR to amplify 52 target regions |
| Sample Types | purified mitochondrial genome to avoid results from nuclear genome |
| Items | N109V0P1, N109V0P2, N109V0P3, N109V0P4. N109V0P5. N109V0P6, N109V0P7, N109V0P8 |
| N109V0 Methylation Panel – Multiplex PCR | ||||||||||||
| PCR ID | Assay | Gene | Coordinates (hg38) | # CpGs | Size (bp) | RSQ | Est. Cov. (S264) | |||||
| N109P1 | ADS5976 | CO1 | ChrM:6445-6540 | 4 | 145 | 0.982 | 1364 | |||||
| ADS5972 | ND1 | ChrM:4126-4152 | 4 | 113 | 0.969 | 579 | ||||||
| TA = 53 | ADS5980 | CYB | ChrM:15349-15436 | 5 | 138 | 0.932 | 3538 | |||||
| ADS9413 | 3′ downstream, CYB | ChrM:16411-16495 | 5 | 141 | 0.891 | 1459 | ||||||
| ADS5987 | CO1 | ChrM:7196-7235 | 6 | 96 | 0.938 | 620 | ||||||
| ADS5997 | 3′ downstream, CYB | ChrM:16328-16360 | 2 | 110 | 0.883 | 516 | ||||||
| ADS5996 | ATP8/ATP6 | ChrM:8532-8583 | 4 | 146 | 0.973 | 173 | ||||||
| ADS5995 | ATP6 | ChrM:8647-8781 | 3 | 200 | 0.976 | 905 | ||||||
| N109P2 | ADS5971 | Intergenic | ChrM:4375-4463 | 4 | 172 | 0.97 | 842 | |||||
| ADS9416 | RNR1 | ChrM:1473-1561 | 6 | 152 | 0.881 | 60 | ||||||
| ADS9515 | ND1 | ChrM:3351-3459 | 8 | 168 | 0.936 | 112 | ||||||
| TA = 55 | ADS5988 | D-Loop | ChrM:7756-7858 | 7 | 193 | 0.954 | 87 | |||||
| ADS5957 | ND5 | ChrM:12862-12888 | 5 | 101 | 0.881 | 601 | ||||||
| N109P3 | ADS5974 | ND1 | ChrM:3642-3706 | 5 | 139 | 0.94 | 1061 | |||||
| TA = 51 | ADS5975 | CO1 | ChrM:6164-6189 | 4 | 90 | 0.948 | 324 | |||||
| ADS5984 | ATP6 | ChrM:9138-9195 | 4 | 140 | 0.947 | 351 | ||||||
| ADS5963 | ND3 | ChrM:10142-10202 | 7 | 149 | 0.959 | 1036 | ||||||
| ADS2886 | CYB | ChrM:14920-15004 | 8 | 150 | 0.956 | 576 | ||||||
| ADS5953 | CYB | ChrM:15499-15616 | 6 | 178 | 0.97 | 213 | ||||||
| ADS5985 | ATP6 | ChrM:8958-9054 | 5 | 166 | 0.987 | 414 | ||||||
| ADS5966 | CO3 | ChrM:9752-9827 | 6 | 136 | 0.917 | 957 | ||||||
| ADS5986 | ATP6 | ChrM:8855-8878 | 2 | 161 | 0.964 | 44 | ||||||
| N109P4 | ADS5961 | ND4 | ChrM:11162-11191 | 3 | 116 | 0.96 | 998 | |||||
| TA = 55 | ADS5994 | CO2 | ChrM:7599-7663 | 3 | 157 | 0.933 | 68 | |||||
| ADS5956 | ND5 | ChrM:13179 | 4 | 121 | 0.868 | 781 | ||||||
| ADS5955 | ND5 | ChrM:13524-13610 | 6 | 155 | 0.938 | 2680 | ||||||
| N109P5 | ADS9500 | RNR1 | ChrM:624-785 | 5 | 246 | 0.869 | 70 | |||||
| ADS5983 | ND5 | ChrM:12735-12817 | 5 | 194 | 0.885 | 783 | ||||||
| TA = 51 | ADS5970 | ND2 | ChrM:4620-4712 | 4 | 169 | 0.972 | 889 | |||||
| ADS9417 | RNR2 | ChrM:1905-2001 | 5 | 155 | 0.851 | 69 | ||||||
| ADS5978 | CO1 | ChrM:7418-7462 | 3 | 124 | 0.854 | 105 | ||||||
| ADS5962 | ND4L | ChrM:10142-10774 | 5 | 190 | 0.887 | 579 | ||||||
| ADS5992 | ND5 | ChrM:13702-13809 | 5 | 170 | 0.861 | 515 | ||||||
| N109P6 | ADS9427 | CO2 | ChrM:8113-8164 | 6 | 130 | 0.904 | 63 | |||||
| TA = 60 | ADS5989 | CO3 | ChrM:9574-9620 | 3 | 130 | 0.963 | 382 | |||||
| ADS5991 | ND5 | ChrM:13287-13405 | 4 | 204 | 0.893 | 2425 | ||||||
| ADS5965 | CO3 | ChrM:9911-10013 | 4 | 166 | 0.925 | 3764 | ||||||
| ADS2887 | ND6 | ChrM:14159-14267 | 4 | 183 | 0.849 | 204 | ||||||
| ADS5959 | ND4 | ChrM:11647-11777 | 8 | 191 | 0.978 | 984 | ||||||
| N109P7 | ADS9415 | RNR1 | ChrM:1177-1322 | 7 | 217 | 0.864 | 174 | |||||
| ADS5969 | ND2 | ChrM:5053-5163 | 4 | 203 | 0.97 | 382 | ||||||
| TA = 51 | ADS5968 | ND2 | ChrM:5236-5399 | 5 | 236 | 0.959 | 242 | |||||
| ADS5960 | ND4 | ChrM:11389-11492 | 6 | 204 | 0.932 | 70 | ||||||
| ADS5958 | ND4 | ChrM:12053-12206 | 5 | 210 | 0.884 | 158 | ||||||
| N109P8 | ADS9419 | RNR2 | ChrM:2642-2718 | 4 | 129 | 0.92 | 2291 | |||||
| ADS5967 | ND2 | ChrM:5459-5470 | 2 | 130 | 0.957 | 1378 | ||||||
| TA = 55 | ADS5954 | ND5 | ChrM:13914-13967 | 3 | 134 | 0.925 | 1562 | |||||
| ADS2921A | ND6 | ChrM:14383-14458 | 2 | 127 | 0.952 | 569 | ||||||
| ADS9424 | CO1 | ChrM:6053-6128 | 4 | 132 | 0.842 | 41 | ||||||
| ADS5979 | CO1 | ChrM:6617-6689 | 3 | 144 | 0.949 | 249 | ||||||
| ADS5964 | D-Loop | ChrM:61-120 | 7 | 114 | 0.863 | 1360 | ||||||
| ADS9425 | CO1 | ChrM:6998-7019 | 4 | 127 | 0.901 | 191 | ||||||
| 8 PCRs | 11 genes | 52 assays | 243 CpGs | 8092 bp | ||||||||
Imprinted Gene Methylation Panel
Genomic imprinting is an epigenetic process that causes genes to express in a way that is particular to the parent of origin. In mammalian development, genomic imprinting, which results in parent-of-origin specific gene expression, is crucial. To assess the maintenance of imprinted gene expression, we used allele-specific RNA-seq on isogenic B6D2F1 mice to assay imprinted genes in tissues from early embryonic tissues between E3.5 to E7.25 and in pluripotent cell lines. Embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs) produced from fertilized embryos and embryos obtained via nuclear transfer (NT) or parthenogenetic activation are included in the cell lines (PGA).

Gene Specific Methylation Panel
Various approaches, such as methylation-sensitive restriction enzymes or bisulfite-modified DNA conversion, can be used to examine DNA methylation at specific genomic regions. Under the right circumstances, the latter mechanism transforms unmethylated cytosines into uracils. DNA methylation (both global and gene-specific) has been identified as an epigenetic process that may have a role in the development of type 2 diabetes mellitus (T2DM). Furthermore, epigenetic therapies have been proposed as a future treatment option for T2DM. Epigenetic modifications demonstrate the disease’s relationship to the environment. Because some epigenetic alterations can be reverted, they could be exploited as therapeutic targets in the future.