EpigenDx analyzes methylation by direct Pyrosequencing after bisulfite conversion of DNA. All of our assays are pre-validated to ensure there is no preferential amplification for either methylated or unmethylated DNA, allowing us to provide you with the highest quality data and results.
Line-1 Global Methylation Analysis
Service Description
Clinical Applications of Line-1 Methylation Analysis
- Drug Screening
- Pre-clinical dosing
- Diagnosis and Prognosis
- PD/PK Studies
- Clinical dose escalating
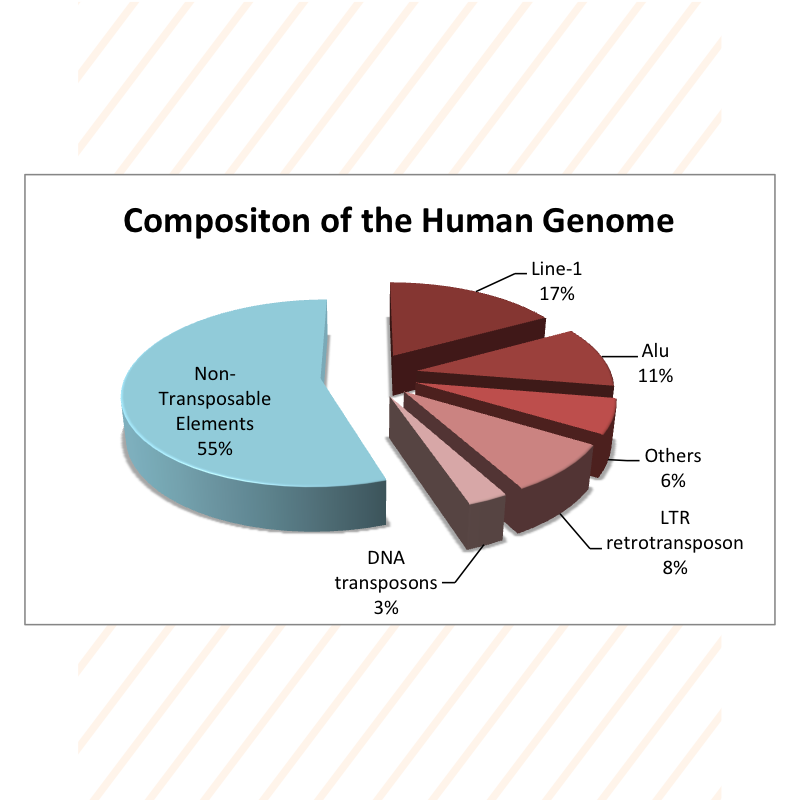
Global DNA Methylation Markers
select a marker, method & get your results
» Human Global Methylation
- → Human LINE-1 Element
- → Human Alu Elements
- → Human SAT2 Element
- → Human HERVE LTR Elements
» Mouse Global Methylation
- → Mouse LINE-1 Element
- → Mouse B1 Element
- → Mouse IAP Element
» Rat Global Methylation
- → Rat LINE-1 Element
- → Rat ID Element
- → Rat Satellite-I
Methods
Human LINE-1 methylation assay (Assay ID: ASY3201) amplifies a 146 base pair (bp) fragment located at the 5’UTR region of the LINE-1 elements (Figure 2). The LINE-1 methylation level can be quantified by Pyrosequencing, MS-HRM, and targeted bisulfite NGS (tNGBS).
These three methods use the same bisulfite PCR but measure methylation levels in different ways:
Pyrosequencing
MS-HRM
Targeted Bisulfite NGS
- LINE-1 Pyrosequencing Assay: Sequence a 36-bp fragment to quantify methylation level of four CpG sites
- MS-HRM Methylation Assay: Provide an average methylation level of the 146-bp region using real-time PCR
- Targeted Bisulfite NGS (tNGBS): Sequence the entire 146-bp amplicon and quantify the methylation of 10 CpG sites with greater than 500x coverage
Validation
Assay Specificity – The LINE-1 Assay (ASY3201) amplifies a conserved LINE-1 element throughout the genome after sodium bisulfite modification, with a PCR amplicon size of 146 bp. It is located at the 5’UTR region of the LINE-1 elements (Figure 2). Pyrosequencing and tNGBS assays demonstrated specificity to the LINE-1 repetitive element sequence.
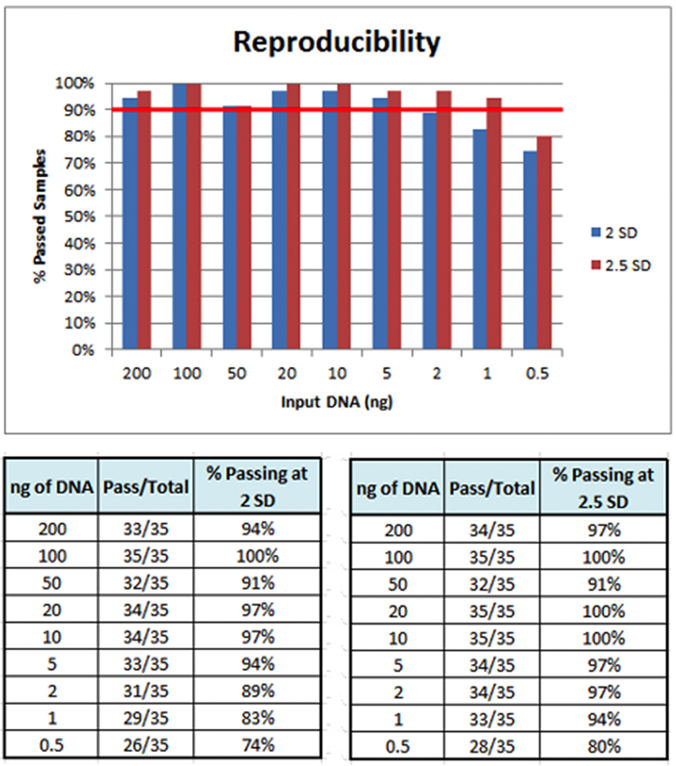
Assay Sensitivity – Assay sensitivity was analyzed by determining the limits of detection (LOD) and quantification (LOO). Both LOD and LOO were set at 1 ng of input DNA.
Line-1 Assay Precision & Accuracy
Assay Precision – The assay was tested for reproducibility on the same day, same operator, and same instrument
(intra-assay precision), as well as reproducibility between different instruments and operators (intermediate precision). The assay was shown to be precise down to 1 ng of input DNA.
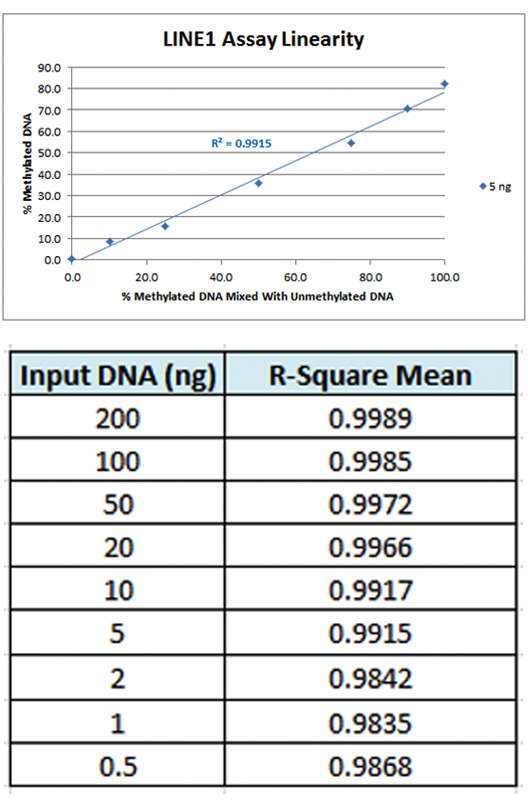
Assay Accuracy – The accuracy of the LINE-1 assay was determined by comparing the methylation data obtained from the validation test samples to the expected values of the control DNA with known methylation levels. An
R-square value of greater than 0.9 at the LOO indicates that the assay is linear and free of PCR Bias. The average R-square for each input DNA amount was calculated and the results are shown in Figure 2. All seven input amounts, down to as low as 0.5 ng of starting DNA, show R-square values above 0.9. An example of the 5 ng input DNA amount is also shown in the figure.
LINE-1 In Cancer
A Biomarker in Cancer Research
LINE-1 hypomethylation has been found in various cancer types, such as bladder cancers4, liver cancer5, and gastrointestinal cancers6. LINE-1 methylation loss can be used as a surrogate marker for cancer diagnosis and prognosis. Genomic DNA isolated from whole blood, buffy coat, buccal cells, serum, FFPE tissue, and other biological specimens is suitable for quantifying LINE-1 methylation level using targeted bisulfite NGS, Pyrosequencing, or MS-HRM.
High-Throughput Drug Screening
Hypomethylating agents (HMAs) such as azacitidine and decitabine are FDA-approved for use in the United States in myelodysplastic syndrome and are being investigated for use in a number of tumors. The Line-1 Pyrosequencing methylation assay has been used for drug screening of compounds that influence global DNA methylation states.
Figure 2 shows the drug-induced Line-1 methylation change at different treatment time points at a selected dose.
A Surrogate Pharmacodynamic (PD) Marker
Line-1 Pyrosequencing methylation analysis has been used as a pharmacodynamic (PD) marker for several clinical trials in cancer drug development. Line-1 methylation can be used as a predictor of clinical response. The time course of LINE-1 methylation following drug administration may be related to HMA concentration in blood or plasma. Simulated LINE-1 matrices can develop a PK/dose-response model which can be used to optimize the dose regimen and to evaluate the dynamic of HMAs.
Looking for a Custom Assay?
No problem, we’ve got you covered. EpigenDx provides custom assay design and analytical services with an assay database of thousands of Off-the-Shelf designs ready to go. Not only do we provide aggressive timelines with regular project updates, you will also get the highest quality technical support and consultation from design phase through project completion and data delivery.
EpigenDx
Experts in DNA Methylation